ATC Code: Not Assigned
Wikipedia: Ospemifene
On February 26, FDA approved Ospemifene (Trade Name: OSPHENA, PubChem: CID 3036505, ChEMBL: CHEMBL2105395, ChemSpider: 2300501) for the treatment of moderate to severe Dyspareunia - symptom of vulvular and vaginal atrophy due to menopause.
Dyspareunia, is pain during or after sexual intercourse. It can affect men, but is significantly more common in women, affecting up to one-fifth of women at some point in their lives. Women with dyspareunia may have pain in the vagina, clitoris or labia. This may be due to medical or psychological causes. There are numerous medical causes of Dyspareunia, like : Vaginismus, Pelvic Inflammatory Disease, Genital or Pelvic Tumors, Urethritis, Urinary Tract Infection, Vaginal Atrophy, Vaginal Dryness, Vulvar Cancer, Childbirth Trauma (postpartum), Skin Conditions (Lichen Sclerosus, Lichen Planus, Eczema, Psoriasis), Female Genital Mutilation, Endometriosis - many of which can be treatable.
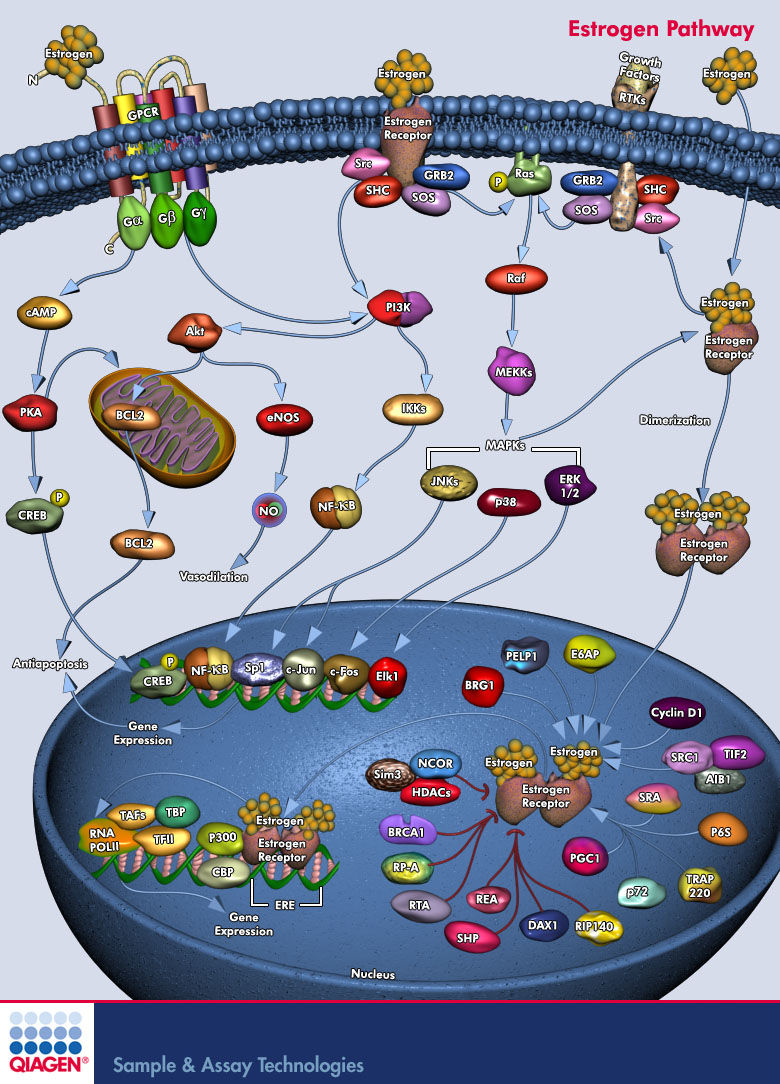
Ospemifene is a novel selective estrogen receptor modulator (SERM) - class of compounds that acts on Estrogen Receptors (ER's). SERM's has a distinguishing characteristic that makes them different from pure receptor agonists and antagonists, which is - that their mode of action is different in various tissues, thereby granting the possibility to selectively inhibit or stimulate estrogen-like action in various tissues (Pub-Med). Ospemifene is an Estrogen agonist/antagonist with tissue selective effects. Its biological actions are mediated through binding to Estrogen Receptors (Short Name: ER, ESR; UniProt: Q92731 and P03372; ChEMBL: CHEMBL2093866). This binding results in activation of estrogenic pathways in some tissues (agonism) and blockade of estrogenic pathways in others (antagonism).
Mechanism of action of SERM's is of mixed agonism/antagonism which may differ depending on the chemical structure, but, for at least for some SERM's, it appears to be related to -
1. The ratio of co-activator to co-repressor proteins in different cell types.
2. The conformation of the estrogen receptor induced by drug binding, which in turn determines how strongly the drug/receptor complex recruits co-activators (resulting in an agonist response) relative to co-repressors (resulting in antagonism).
The protein sequences of human ER-alpha (ESR1) and ER-beta (ESR2) can be downloaded in fasta format from the link here. (courtesy UniProt)
Compound Name : Z-2-[4-(4-chloro-1,2-diphenylbut-1-enyl)phenoxy]ethanol
Canonical SMILES : OCCOc1ccc(cc1)\C(=C(\CCCl)/c2ccccc2)\c3ccccc3
Standard InChI : InChI=1S/C24H23ClO2/c25-16-15-23(19-7-3-1-4-8-19)24(20-9-5-2-6-10-20)21-11-13-22(14-12-21)27-18-17-26/h1-14,26H,15-18H2/b24-23-
Standard InChI Key : LUMKNAVTFCDUIE-VHXPQNKSSA-N
Ospemifene, an ER agonist/antagonist, has a molecular weight of 378.9. The recommended dosage is 60 mg, available in the form of table for oral administration. After single dosage of Ospemifene under fasted conditions, mean Cmax and AUC (0 to infinity) were 533 ng/mL and 4165 ng.hr/mL, respectively. With a high fat/high diet, mean Cmax and AUC (0 to infinity) were 1198 ng/mL and 7521 ng.hr/mL, respectively. It is highly bound to serum proteins ( >99%) with apparent volume of distribution of 448 L. Ospemifene primarily undergoes metabolism via CYP3A4, CYP2C9 and CYP2C19 and the major metabolite was 4-hydroxyospemifene. Clearance of ospemifene was 9.16 L/hr and terminal half-life was 26 hrs. Following an oral administration of ospemifene, approximately 75% and 7% of the dose was excreted in feces and urine, respectively.
Osphena comes with a boxed warning in the form of Endometrial Cancer and Cardiovascular Disorders. Since Ospemifene is an ER agonist/antagonist with tissue selective effects. In endometrium, due to Ospemifene agonistic effects, there is an increased risk of endometrial cancer in a woman with a uterus who uses unopposed estrogens. Adding a progestin to estrogen therapy reduces the risk of endometrial hyperplasia, which may be a precursor to endometrial cancer. There is a reported increased risk of stroke and deep vein thrombosis in postmenopausal women who received daily oral conjugated estrogens alone therapy over 7.1 years. So, Ospemifene should be prescribed for the shortest duration consistent with treatment goals.
Full prescribing information can be found here.
The license holder is Shionogi Inc., and the product website is www.osphena.com.





Comments