ATC Code:
Wikipedia: Raxibacumab
On December 14th 2012 the FDA approved Raxibacumab for the treatment of inhalation anthrax, a form of anthrax caused by the inhalation of anthrax spores. The drug is also approved to treat inhalation anthrax when alternative therapies are not available or appropriate. Raxibacumab is a 146 kDa monoclonal antibody that is designed to neutralize the toxin secreted by Bacillus Anthracis. The FDA granted raxibacumab fast track designation, priority review, and orphan product designation.
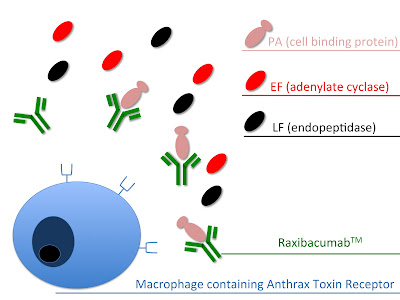
Bacillus Anthracis toxin (Anthrax toxin) is a secreted three protein exotoxin. It consists of two enzyme components; lethal factor (LF, PDB 1PWU), a bacterial endopeptidase and edema factor (EF, PDB 1PWW), a bacterial adenylate cyclase. These are combined with one cell-binding protein; protective antigen (PA, PDB 1ACC). The individual components are non toxic and the combination of the enzyme components with the cell-binding protein makes them toxic. PA, in the form of a 83kDa protein, binds to the Anthrax Toxin receptor. Upon binding a 20kDa fragment is cleaved of the protein. The remaining protein (PA63) self assembles into a ring shaped oligomer. This oligomer acts as a pore precursor through which the enzymatic components enter the cell. EF, an 88kDa protein, acts as a Ca2+ and calmodulin dependant adenylate cyclase, raising cAMP levels (up to 200 fold in CHO cells) and disturbing water homeostasis in the cell. In turn disturbing signaling pathways and immune function. LF, an 89kDa protein, is a Zn2+ dependant endopeptidase. The protein cleaves mitogen-activated proten kinase kinases (MAPKKs). This leads to altered signalling pathways and apoptosis.
Raxibacumab, efficacy has not been tested in humans but instead in monkey's and rabbit's for ethical reasons. Safety trials were conducted in 326 healthy human volunteers.
Raxibacumab is available as a single-use vial which contains 1700 mg/34 mL (50 mg/mL) raxibacumab injection. Raxibacumab is administered as a single dose of 40 mg/kg intravenously over 2 hours and 15 minutes after dilution in 0.9% Sodium Chloride Injection, USP (normal saline) to a final volume of 250 mL.
The PK of raxibacumab are linear over the dose range of 1 to 40 mg/kg following single IV dosing in humans. Following single IV administration of raxibacumab 40 mg/kg in healthy, male and female human subjects, the mean Cmax and AUCinf were 1020.3 ± 140.6 mcg/mL and 15845.8 ± 4333.5 mcg·day/mL, respectively. Mean raxibacumab steady-state volume of distribution was greater than plasma volume, suggesting some tissue distribution. Clearance values were much smaller than the glomerular filtration rate indicating that there is virtually no renal clearance of raxibacumab.
The license holder is GlaxoSmithKline and the prescribing information can be found here.
Comments